
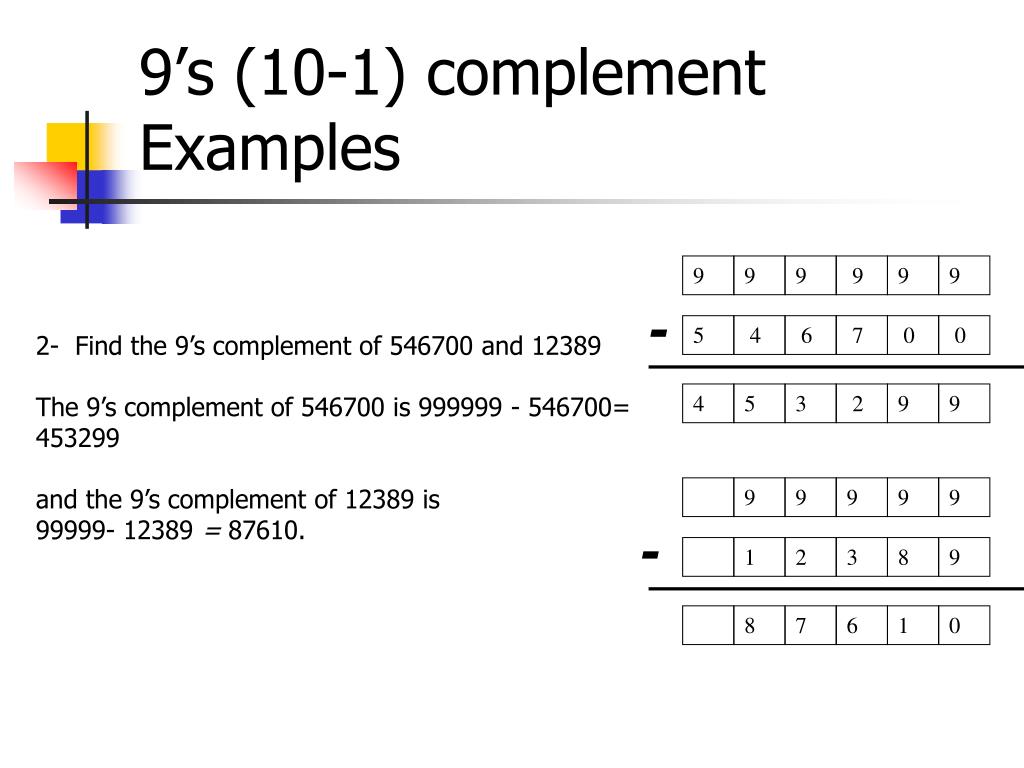
When designing and synthesizing synthetic DNA oligonucleotides, for example for use in antisense therapy or as PCR probes, it is often desirable to either increase or decrease the stability of the duplex formed on binding to complementary DNA. However, this formula only applies to DNA duplexes containing exclusively Watson-Crick A.T and G.C base pairs under a well-defined set of conditions, and cannot be used to determine the melting temperatures of mismatch-containing duplexes.
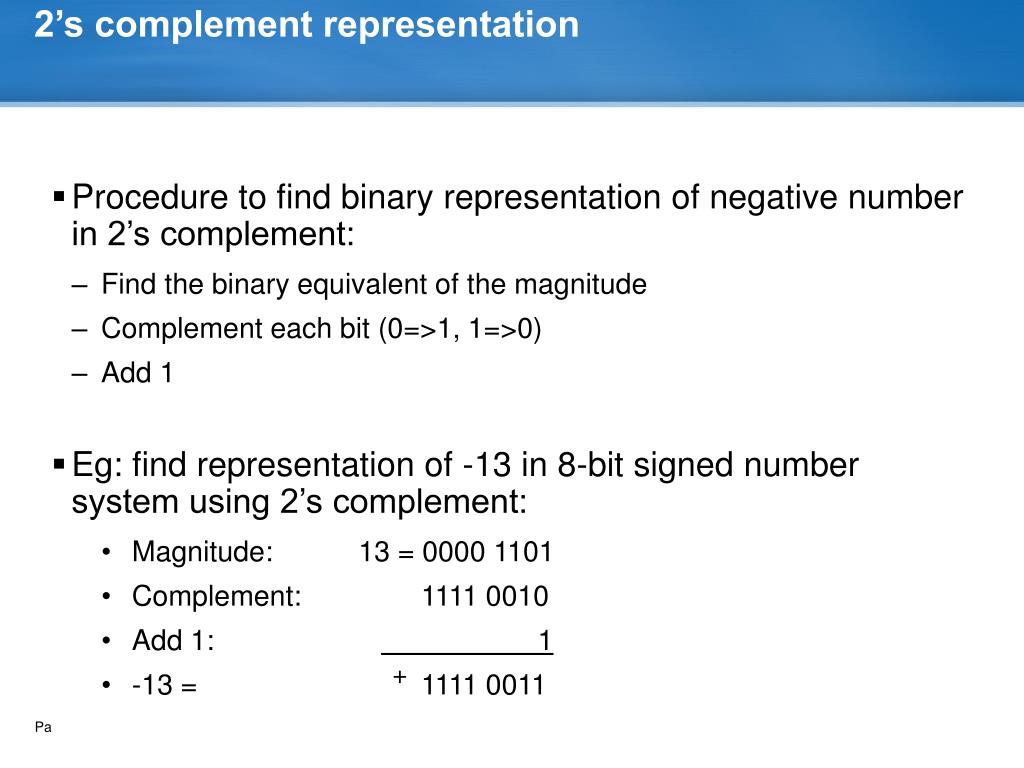
\footnotesize\begin n i is the number of bases of type i \footnotesize i i present. However, it is probably more informative to consider base pairs rather than individual bases as discrete units in order to visualize the stabilizing effects of base stacking. Base-stacking interactions in nucleic acid duplexes are partly inter-strand and partly intra-strand in nature. Base-stacking interactions are hydrophobic and electrostatic in nature, and depend on the aromaticity of the bases and their dipole moments. The individual bases form strong stacking interactions which are major contributors to duplex stability, as base stacking is much more prevalent in duplexes than in single strands ( Figure 1). Inter-strand hydrogen bonding is clearly important in driving the formation of DNA duplexes, but it is by no means the only contributing factor. Not surprisingly the coming together of two large oligomeric molecules is entropically unfavourable Δ S is negative). For duplexes of any significant length this is an exothermic process at ambient temperature. The overall process is one of "hydrogen bond exchange" and the net change in enthalpy upon duplex formation is partly due to Δ H (H-bonds formed) − Δ H (H-bonds broken). Some of these bonds must be broken during duplex formation as the inter-base hydrogen bonds are formed. The heterocyclic bases of single-stranded DNA have polar amino, amidino, guanidino and carbonyl groups that form a complex network of hydrogen bonds with the surrounding water molecules. Factors influencing DNA duplex stabilityĭNA duplex stability is determined primarily by hydrogen bonding, but base stacking also plays an important role. To gain an insight into DNA duplex stability, and how it is affected by changes in primary structure, scientists have studied the structure and thermodynamic stability of a variety of DNA duplexes by using a combination of physical methods including X-ray crystallography, ultraviolet (UV) melting and NMR.
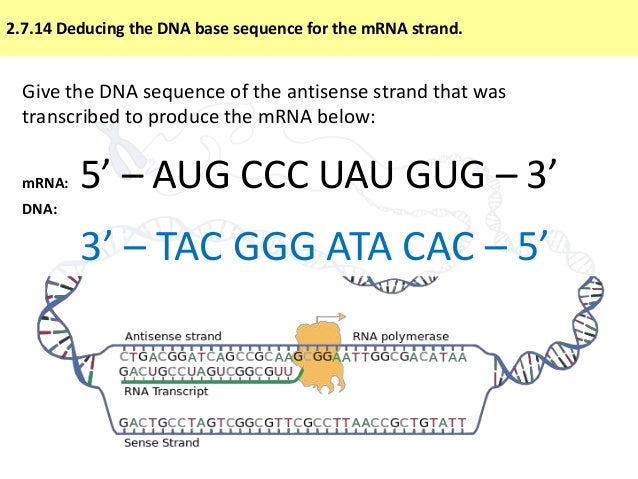
This instability is exploited by proofreading enzymes which recognize the mutation and replace it with the correct nucleotide (see Mutagenesis and DNA repair). For example, mutations in the base sequence that result from errors that occur during DNA replication can result in mismatches that lead to relatively unstable duplexes. Slight variations in the DNA sequence can have profound implications on the stability of the DNA duplex. The stability of the DNA double helix depends on a fine balance of interactions including hydrogen bonds between bases, hydrogen bonds between bases and surrounding water molecules, and base-stacking interactions between adjacent bases.


 0 kommentar(er)
0 kommentar(er)
